Filters
Host (767983)
Bovine (1070)Canine (20)Cat (401)Chicken (1579)Cod (2)Cow (333)Crab (15)Dog (524)Dolphin (2)Duck (13)E Coli (239151)Equine (7)Feline (1785)Ferret (306)Fish (125)Frog (52)Goat (36654)Guinea Pig (732)Hamster (1376)Horse (903)Insect (2052)Mammalian (512)Mice (6)Monkey (624)Mouse (96348)Pig (197)Porcine (70)Rabbit (358350)Rat (11727)Ray (55)Salamander (4)Salmon (15)Shark (3)Sheep (4248)Snake (4)Swine (301)Turkey (57)Whale (3)Yeast (5335)Zebrafish (3022)Isotype (153116)
IgA (13004)IgA1 (819)IgA2 (298)IgD (1957)IgE (5308)IgG (85179)IgG1 (16541)IgG2 (1228)IgG3 (2593)IgG4 (1509)IgM (21994)IgY (2686)Label (240992)
AF488 (2882)AF594 (726)AF647 (2718)ALEXA (12103)ALEXA FLUOR 350 (298)ALEXA FLUOR 405 (303)ALEXA FLUOR 488 (715)ALEXA FLUOR 532 (303)ALEXA FLUOR 555 (317)ALEXA FLUOR 568 (296)ALEXA FLUOR 594 (342)ALEXA FLUOR 633 (305)ALEXA FLUOR 647 (650)ALEXA FLUOR 660 (295)ALEXA FLUOR 680 (465)ALEXA FLUOR 700 (2)ALEXA FLUOR 750 (457)ALEXA FLUOR 790 (258)Alkaline Phosphatase (813)Allophycocyanin (22)ALP (387)AMCA (133)AP (1211)APC (14715)APC C750 (13)Apc Cy7 (1248)ATTO 390 (3)ATTO 488 (6)ATTO 550 (1)ATTO 594 (5)ATTO 647N (4)AVI (53)Beads (234)Beta Gal (2)BgG (1)BIMA (6)Biotin (27795)Biotinylated (1810)Blue (708)BSA (876)BTG (46)C Terminal (688)CF Blue (19)Colloidal (22)Conjugated (29303)Cy (160)Cy3 (449)Cy5 (2100)Cy5 5 (2520)Cy5 PE (1)Cy7 (3697)Dual (170)DY549 (3)DY649 (3)Dye (1)DyLight (1301)DyLight 405 (11)DyLight 488 (222)DyLight 549 (22)DyLight 594 (88)DyLight 649 (4)DyLight 650 (35)DyLight 680 (20)DyLight 800 (24)Fam (13)Fc Tag (8)FITC (29583)Flag (213)Fluorescent (146)GFP (561)GFP Tag (178)Glucose Oxidase (59)Gold (511)Green (580)GST (713)GST Tag (324)HA Tag (453)His (642)His Tag (520)Horseradish (550)HRP (12962)HSA (247)iFluor (16571)Isoform b (31)KLH (87)Luciferase (102)Magnetic (259)MBP (341)MBP Tag (93)Myc Tag (424)OC 515 (1)Orange (78)OVA (99)Pacific Blue (213)Particle (64)PE (34110)PerCP (7934)Peroxidase (1361)POD (11)Poly Hrp (94)Poly Hrp40 (13)Poly Hrp80 (3)Puro (32)Red (2490)RFP Tag (63)Rhodamine (614)RPE (910)S Tag (194)SCF (182)SPRD (351)Streptavidin (55)SureLight (77)T7 Tag (97)Tag (4845)Texas (1300)Texas Red (1282)Triple (10)TRITC (1454)TRX tag (90)Unconjugated (2102)Unlabeled (218)Yellow (84)Pathogen (488409)
Adenovirus (8498)AIV (317)Bordetella (25037)Borrelia (18291)Candida (17805)Chikungunya (590)Chlamydia (17582)CMV (121404)Coronavirus (5969)Coxsackie (749)Dengue (2877)EBV (1485)Echovirus (215)Enterovirus (677)Hantavirus (260)HAV (906)HBV (2045)HHV (838)HIV (7781)hMPV (277)HSV (2328)HTLV (641)Influenza (21967)Isolate (1208)KSHV (396)Lentivirus (3754)Lineage (2912)Lysate (127755)Marek (95)Measles (1172)Parainfluenza (1692)Poliovirus (2680)Poxvirus (82)Rabies (1527)Reovirus (538)Retrovirus (1070)Rhinovirus (512)Rotavirus (5356)RSV (1754)Rubella (1070)SIV (279)Strain (67807)Vaccinia (7182)VZV (659)WNV (370)Species (2793042)
Alligator (10)Bovine (150318)Canine (112024)Cat (13062)Chicken (104944)Cod (1)Cow (2029)Dog (11449)Dolphin (21)Duck (9739)Equine (2060)Feline (1004)Ferret (259)Fish (12830)Frog (1)Goat (81348)Guinea Pig (78672)Hamster (36716)Horse (40600)Human (900649)Insect (651)Lemur (110)Lizard (24)Monkey (102126)Mouse (444461)Pig (24064)Porcine (123365)Rabbit (117244)Rat (325094)Ray (401)Salmon (337)Seal (8)Shark (29)Sheep (96001)Snake (11)Swine (493)Toad (4)Turkey (244)Turtle (75)Whale (45)Zebrafish (519)Technique (10953826)
Activation (208929)Activity (14692)Affinity (88781)Agarose (3192)Aggregation (204)Antigen (163775)Apoptosis (25807)Array (3334)Blocking (105075)Blood (9047)Blot (15164)ChiP (1064)Chromatin (5450)Colorimetric (15875)Control (36297)Culture (5608)Cytometry (5665)Depletion (139)DNA (176413)Dot (315)EIA (1076)Electron (6114)Electrophoresis (442)ELISA (2987962)Elispot (2324)Enzymes (52676)Exosome (4709)Extract (1502)Fab (3121)FACS (61)FC (97132)Filter Tips (388)Flow (14143)Fluorometric (1620)Formalin (122)Frozen (22695)Functional (568)Gel (3290)HTS (167)IHC (27111)Immunoassay (8460)Immunofluorescence (5625)Immunohistochemistry (81)Immunoprecipitation (80)intracellular (5387)IP (4346)iPSC (342)Isotype (17708)Lateral (1686)Lenti (224152)Light (42142)Microarray (203)MicroRNA (4889)Microscopy (811)miRNA (117341)Monoclonal (1425181)Multi (4676)Multiplex (7715)Negative (4333)PAGE (2638)Panel (2360)Paraffin (2734)PBS (27857)PCR (9)Peptide (329762)PerCP (68483)Polyclonal (3439718)Positive (6592)Precipitation (84)Premix (162)Primers (47329)Probe (3204)Profile (561)Pure (10549)Purification (15)Purified (111201)Real Time (3368)Resin (3255)Reverse (2876)RIA (523)RNAi (63118)Rox (1305)RT PCR (6833)Sample (4708)SDS (1876)Section (2971)Separation (245)Sequencing (224)Shift (13)siRNA (614632)Standard (82523)Sterile (12102)Strip (2488)Taq (2)Tip (1730)Tissue (80783)Tube (4389)Vitro (3590)Vivo (2367)WB (2998)Western Blot (14442)Tissue (1939321)
Adenocarcinoma (1075)Adipose (3101)Adrenal (645)Adult (4585)Amniotic (65)Animal (2573)Aorta (436)Appendix (89)Array (2022)Ascites (4737)Bile Duct (20)Bladder (1636)Blood (8238)Bone (23970)Brain (27644)Breast (10082)Calvaria (28)Carcinoma (12693)cDNA (58563)Cell (397549)Cellular (8128)Cerebellum (700)Cervix (232)Child (1)Choroid (19)Colon (3755)Connective (2945)Contaminant (3)Control (80450)Cord (661)Corpus (148)Cortex (686)Dendritic (1821)Diseased (265)Donor (1361)Duct (763)Duodenum (641)Embryo (424)Embryonic (4306)Endometrium (454)Endothelium (1122)Epidermis (166)Epithelium (3596)Esophagus (670)Exosome (4229)Eye (1882)Female (476)Frozen (2671)Gallbladder (155)Genital (5)Gland (3286)Granulocyte (8035)Heart (6154)Hela (413)Hippocampus (325)Histiocytic (73)Ileum (201)Insect (4882)Intestine (1945)Isolate (1208)Jejunum (175)Kidney (7357)Langerhans (282)Leukemia (20040)Liver (15485)Lobe (812)Lung (5869)Lymph (1208)Lymphatic (632)lymphocyte (20657)Lymphoma (11533)Lysate (127755)Lysosome (2774)Macrophage (28125)Male (1603)Malignant (1387)Mammary (1905)Mantle (1041)Marrow (2018)Mastocytoma (3)Matched (11710)Medulla (156)Melanoma (14602)Membrane (103016)Metastatic (3023)Mitochondrial (153311)Muscle (33392)Myeloma (748)Myocardium (11)Nerve (5919)Neuronal (15439)Node (1206)Normal (8945)Omentum (10)Ovarian (2328)Ovary (1157)Pair (47181)Pancreas (2658)Panel (1636)Penis (64)Peripheral (1785)Pharynx (120)Pituitary (5073)Placenta (3784)Prostate (9194)Proximal (315)Rectum (316)Region (200946)Retina (956)Salivary (2813)Sarcoma (6636)Section (2895)Serum (23693)Set (167349)Skeletal (12077)Skin (1755)Smooth (7028)Spinal (424)Spleen (2163)Stem (8361)Stomach (922)Stroma (49)Subcutaneous (47)Testis (14837)Thalamus (127)Thoracic (60)Throat (42)Thymus (2673)Thyroid (13057)Tongue (144)Total (9944)Trachea (227)Transformed (175)Tubule (48)Tumor (70119)Umbilical (208)Ureter (73)Urinary (2404)Uterine (301)The Difference Between MIT “Microcompartments” and Bacterial Microcompartments
The word microcompartment sounds universal. It suggests a small, functional sub-unit within a biological system. But in modern biology, it describes two completely different entities.
In 2025, researchers at the Massachusetts Institute of Technology (MIT) reported the discovery of microcompartments in the 3D genome, tiny DNA loops that survive through cell division. These eukaryotic microcompartments exist inside the nucleus and shape how the genome folds and regulates genes.
Meanwhile, for decades, microbiologists have studied bacterial microcompartments (BMCs), protein-based organelles that package enzymes and control metabolic reactions.
Although the same word is used, these systems have different origins, materials, and purposes. This article compares them in depth.
Genprice
Scientific Publications

The Difference Between MIT “Microcompartments” and Bacterial Microcompartments
Origin and Biological Context

Summary:
MIT’s microcompartments are structural DNA loops in eukaryotes.
Bacterial microcompartments are protein nanostructures enclosing enzymes.
2. Structural Composition
MIT Microcompartments
- Composed of chromatin, which includes DNA, histones, and transcription-related proteins.
- Represent 3D genome loops that bring enhancers and promoters close together.
- Size range: typically a few kilobases of DNA interacting within nanometers to micrometers in space.
- Detected by Region-Capture Micro-C (RC-MC), a next-generation chromosome conformation technique offering base-pair-level resolution.

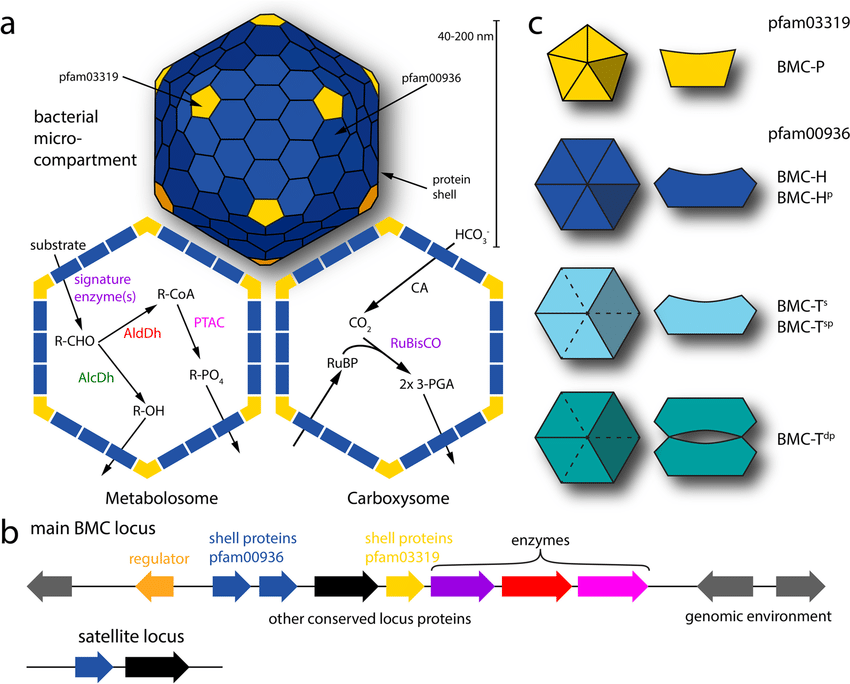
Bacterial Microcompartments
- Composed entirely of proteins forming polyhedral shells.
- Built from hexameric, pentameric, and trimeric shell proteins (BMC-H, BMC-P, BMC-T).
- Inside the shell: enzymes and cofactors for specific metabolic pathways (e.g., carbon fixation, ethanolamine degradation).
- Diameter: 100–200 nm, visible under electron microscopy.
Summary:
Eukaryotic microcompartments are DNA loops.
Bacterial microcompartments are protein cages.
3. Biological Function
MIT Microcompartments : Genome Architecture
- Function as local hubs of regulatory DNA interactions.
- Connect enhancers and promoters, enabling precise transcriptional control.
- Persist through mitosis, meaning some 3D genome structure survives cell division.
- Likely contribute to epigenetic memory and rapid reactivation of genes after division.
- Their formation depends on chromatin compaction and homotypic affinity of regulatory elements.
Bacterial Microcompartments : Metabolic Control
- Function as metabolic organelles in bacteria.
- Sequester specific enzymes to improve reaction efficiency and prevent toxicity.
- Classic examples:
- Carboxysomes (in cyanobacteria) : fix carbon dioxide using RuBisCO and carbonic anhydrase.
- Pdu microcompartments : process 1,2-propanediol.
- Eut microcompartments : metabolize ethanolamine.
- Serve as primitive analogues of organelles, enabling spatial organization in bacteria that lack membrane-bound compartments.
Summary:
Eukaryotic microcompartments regulate gene expression.
Bacterial microcompartments regulate metabolic flux.
4. Key Differences

Conclusion
"Microcompartment" means different things in different biological worlds.
- MIT microcompartments are 3D chromatin loops that organize the eukaryotic genome and persist through mitosis.
- Bacterial microcompartments are protein nanostructures that isolate metabolic reactions.
Both represent elegant examples of biological compartmentalization : one governing the information flow of DNA, the other governing the chemical flow of metabolism.
Together they show that across life’s hierarchy, structure and function are always organized by the same universal principle: compartmentalization for control.
