Filters
Host (767983)
Bovine (1070)Canine (20)Cat (401)Chicken (1579)Cod (2)Cow (333)Crab (15)Dog (524)Dolphin (2)Duck (13)E Coli (239151)Equine (7)Feline (1785)Ferret (306)Fish (125)Frog (52)Goat (36654)Guinea Pig (732)Hamster (1376)Horse (903)Insect (2052)Mammalian (512)Mice (6)Monkey (624)Mouse (96348)Pig (197)Porcine (70)Rabbit (358350)Rat (11727)Ray (55)Salamander (4)Salmon (15)Shark (3)Sheep (4248)Snake (4)Swine (301)Turkey (57)Whale (3)Yeast (5335)Zebrafish (3022)Isotype (153116)
IgA (13004)IgA1 (819)IgA2 (298)IgD (1957)IgE (5308)IgG (85179)IgG1 (16541)IgG2 (1228)IgG3 (2593)IgG4 (1509)IgM (21994)IgY (2686)Label (240992)
AF488 (2882)AF594 (726)AF647 (2718)ALEXA (12103)ALEXA FLUOR 350 (298)ALEXA FLUOR 405 (303)ALEXA FLUOR 488 (715)ALEXA FLUOR 532 (303)ALEXA FLUOR 555 (317)ALEXA FLUOR 568 (296)ALEXA FLUOR 594 (342)ALEXA FLUOR 633 (305)ALEXA FLUOR 647 (650)ALEXA FLUOR 660 (295)ALEXA FLUOR 680 (465)ALEXA FLUOR 700 (2)ALEXA FLUOR 750 (457)ALEXA FLUOR 790 (258)Alkaline Phosphatase (813)Allophycocyanin (22)ALP (387)AMCA (133)AP (1211)APC (14715)APC C750 (13)Apc Cy7 (1248)ATTO 390 (3)ATTO 488 (6)ATTO 550 (1)ATTO 594 (5)ATTO 647N (4)AVI (53)Beads (234)Beta Gal (2)BgG (1)BIMA (6)Biotin (27795)Biotinylated (1810)Blue (708)BSA (876)BTG (46)C Terminal (688)CF Blue (19)Colloidal (22)Conjugated (29303)Cy (160)Cy3 (449)Cy5 (2100)Cy5 5 (2520)Cy5 PE (1)Cy7 (3697)Dual (170)DY549 (3)DY649 (3)Dye (1)DyLight (1301)DyLight 405 (11)DyLight 488 (222)DyLight 549 (22)DyLight 594 (88)DyLight 649 (4)DyLight 650 (35)DyLight 680 (20)DyLight 800 (24)Fam (13)Fc Tag (8)FITC (29583)Flag (213)Fluorescent (146)GFP (561)GFP Tag (178)Glucose Oxidase (59)Gold (511)Green (580)GST (713)GST Tag (324)HA Tag (453)His (642)His Tag (520)Horseradish (550)HRP (12962)HSA (247)iFluor (16571)Isoform b (31)KLH (87)Luciferase (102)Magnetic (259)MBP (341)MBP Tag (93)Myc Tag (424)OC 515 (1)Orange (78)OVA (99)Pacific Blue (213)Particle (64)PE (34110)PerCP (7934)Peroxidase (1361)POD (11)Poly Hrp (94)Poly Hrp40 (13)Poly Hrp80 (3)Puro (32)Red (2490)RFP Tag (63)Rhodamine (614)RPE (910)S Tag (194)SCF (182)SPRD (351)Streptavidin (55)SureLight (77)T7 Tag (97)Tag (4845)Texas (1300)Texas Red (1282)Triple (10)TRITC (1454)TRX tag (90)Unconjugated (2102)Unlabeled (218)Yellow (84)Pathogen (488409)
Adenovirus (8498)AIV (317)Bordetella (25037)Borrelia (18291)Candida (17805)Chikungunya (590)Chlamydia (17582)CMV (121404)Coronavirus (5969)Coxsackie (749)Dengue (2877)EBV (1485)Echovirus (215)Enterovirus (677)Hantavirus (260)HAV (906)HBV (2045)HHV (838)HIV (7781)hMPV (277)HSV (2328)HTLV (641)Influenza (21967)Isolate (1208)KSHV (396)Lentivirus (3754)Lineage (2912)Lysate (127755)Marek (95)Measles (1172)Parainfluenza (1692)Poliovirus (2680)Poxvirus (82)Rabies (1527)Reovirus (538)Retrovirus (1070)Rhinovirus (512)Rotavirus (5356)RSV (1754)Rubella (1070)SIV (279)Strain (67807)Vaccinia (7182)VZV (659)WNV (370)Species (2793042)
Alligator (10)Bovine (150318)Canine (112024)Cat (13062)Chicken (104944)Cod (1)Cow (2029)Dog (11449)Dolphin (21)Duck (9739)Equine (2060)Feline (1004)Ferret (259)Fish (12830)Frog (1)Goat (81348)Guinea Pig (78672)Hamster (36716)Horse (40600)Human (900649)Insect (651)Lemur (110)Lizard (24)Monkey (102126)Mouse (444461)Pig (24064)Porcine (123365)Rabbit (117244)Rat (325094)Ray (401)Salmon (337)Seal (8)Shark (29)Sheep (96001)Snake (11)Swine (493)Toad (4)Turkey (244)Turtle (75)Whale (45)Zebrafish (519)Technique (10953826)
Activation (208929)Activity (14692)Affinity (88781)Agarose (3192)Aggregation (204)Antigen (163775)Apoptosis (25807)Array (3334)Blocking (105075)Blood (9047)Blot (15164)ChiP (1064)Chromatin (5450)Colorimetric (15875)Control (36297)Culture (5608)Cytometry (5665)Depletion (139)DNA (176413)Dot (315)EIA (1076)Electron (6114)Electrophoresis (442)ELISA (2987962)Elispot (2324)Enzymes (52676)Exosome (4709)Extract (1502)Fab (3121)FACS (61)FC (97132)Filter Tips (388)Flow (14143)Fluorometric (1620)Formalin (122)Frozen (22695)Functional (568)Gel (3290)HTS (167)IHC (27111)Immunoassay (8460)Immunofluorescence (5625)Immunohistochemistry (81)Immunoprecipitation (80)intracellular (5387)IP (4346)iPSC (342)Isotype (17708)Lateral (1686)Lenti (224152)Light (42142)Microarray (203)MicroRNA (4889)Microscopy (811)miRNA (117341)Monoclonal (1425181)Multi (4676)Multiplex (7715)Negative (4333)PAGE (2638)Panel (2360)Paraffin (2734)PBS (27857)PCR (9)Peptide (329762)PerCP (68483)Polyclonal (3439718)Positive (6592)Precipitation (84)Premix (162)Primers (47329)Probe (3204)Profile (561)Pure (10549)Purification (15)Purified (111201)Real Time (3368)Resin (3255)Reverse (2876)RIA (523)RNAi (63118)Rox (1305)RT PCR (6833)Sample (4708)SDS (1876)Section (2971)Separation (245)Sequencing (224)Shift (13)siRNA (614632)Standard (82523)Sterile (12102)Strip (2488)Taq (2)Tip (1730)Tissue (80783)Tube (4389)Vitro (3590)Vivo (2367)WB (2998)Western Blot (14442)Tissue (1939321)
Adenocarcinoma (1075)Adipose (3101)Adrenal (645)Adult (4585)Amniotic (65)Animal (2573)Aorta (436)Appendix (89)Array (2022)Ascites (4737)Bile Duct (20)Bladder (1636)Blood (8238)Bone (23970)Brain (27644)Breast (10082)Calvaria (28)Carcinoma (12693)cDNA (58563)Cell (397549)Cellular (8128)Cerebellum (700)Cervix (232)Child (1)Choroid (19)Colon (3755)Connective (2945)Contaminant (3)Control (80450)Cord (661)Corpus (148)Cortex (686)Dendritic (1821)Diseased (265)Donor (1361)Duct (763)Duodenum (641)Embryo (424)Embryonic (4306)Endometrium (454)Endothelium (1122)Epidermis (166)Epithelium (3596)Esophagus (670)Exosome (4229)Eye (1882)Female (476)Frozen (2671)Gallbladder (155)Genital (5)Gland (3286)Granulocyte (8035)Heart (6154)Hela (413)Hippocampus (325)Histiocytic (73)Ileum (201)Insect (4882)Intestine (1945)Isolate (1208)Jejunum (175)Kidney (7357)Langerhans (282)Leukemia (20040)Liver (15485)Lobe (812)Lung (5869)Lymph (1208)Lymphatic (632)lymphocyte (20657)Lymphoma (11533)Lysate (127755)Lysosome (2774)Macrophage (28125)Male (1603)Malignant (1387)Mammary (1905)Mantle (1041)Marrow (2018)Mastocytoma (3)Matched (11710)Medulla (156)Melanoma (14602)Membrane (103016)Metastatic (3023)Mitochondrial (153311)Muscle (33392)Myeloma (748)Myocardium (11)Nerve (5919)Neuronal (15439)Node (1206)Normal (8945)Omentum (10)Ovarian (2328)Ovary (1157)Pair (47181)Pancreas (2658)Panel (1636)Penis (64)Peripheral (1785)Pharynx (120)Pituitary (5073)Placenta (3784)Prostate (9194)Proximal (315)Rectum (316)Region (200946)Retina (956)Salivary (2813)Sarcoma (6636)Section (2895)Serum (23693)Set (167349)Skeletal (12077)Skin (1755)Smooth (7028)Spinal (424)Spleen (2163)Stem (8361)Stomach (922)Stroma (49)Subcutaneous (47)Testis (14837)Thalamus (127)Thoracic (60)Throat (42)Thymus (2673)Thyroid (13057)Tongue (144)Total (9944)Trachea (227)Transformed (175)Tubule (48)Tumor (70119)Umbilical (208)Ureter (73)Urinary (2404)Uterine (301)Revolutionizing Microbial Transcriptomics: A Breakthrough in Bacterial scRNA-seq Protocols
Genprice
Scientific Publications
Revolutionizing Microbial Transcriptomics: A Breakthrough in Bacterial scRNA-seq Protocols
Introduction
In the evolving field of microbial biology, the development of a robust bacterial single-cell RNA sequencing (scRNA-seq) protocol marks a pivotal shift. While scRNA-seq has transformed research in multicellular organisms, its application in prokaryotes, especially bacteria, has remained largely elusive due to intrinsic technical limitations. A newly engineered protocol now allows for detailed transcriptional profiling of individual bacterial cells, opening doors for deep analysis of microbial behavior, adaptability, and gene expression variability.
This innovation provides researchers with a scalable and reliable framework to assess cell-to-cell differences across complex bacterial communities. Whether it’s biofilm formation, metabolic switching, or dormancy responses, this protocol enables the quantification of transcriptomic activity that was previously hidden beneath population averages.
Why Bacterial scRNA-seq Was Technically Difficult Until Now
Applying scRNA-seq to bacteria was a monumental task due to a number of well-documented complications:
- Extremely low mRNA content per bacterial cell compared to eukaryotic cells.
- Absence of polyadenylated tails, which are necessary for most eukaryotic mRNA capture methods.
- Rigid cell walls that hinder effective lysis in droplet- or plate-based workflows.
- Fast degradation of bacterial transcripts, which limits capture efficiency without rapid preservation.
These hurdles made conventional droplet-based and microwell-based scRNA-seq platforms poorly suited for bacterial analysis.
Sources such as ncbi.nlm.nih.gov, genome.gov, and cancer.gov have documented numerous unsuccessful or limited attempts in adapting traditional single-cell methods to bacterial systems, especially for Gram-positive and spore-forming species.
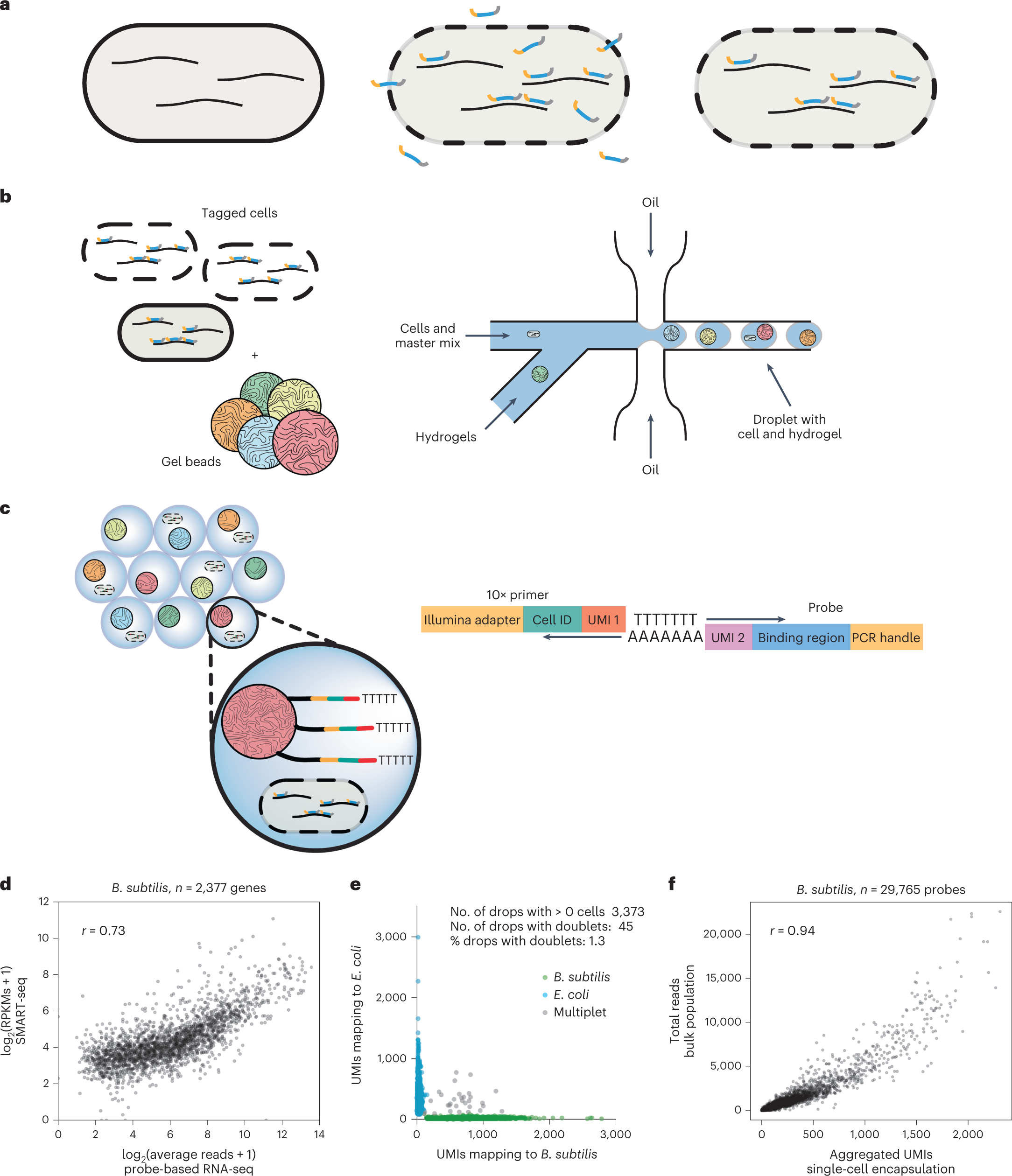
The Protocol: From Bacterial Lysis to Gene Expression Matrix
The new protocol achieves what previous iterations could not — consistent and scalable detection of bacterial transcripts at the single-cell level. Here's how it works, step by step:
🧬 1. Efficient Cell Isolation
Bacteria are isolated using size-optimized microfluidic chips or microwell arrays that can accurately partition submicron cells, unlike standard eukaryotic workflows.
🔓 2. Integrated Enzymatic and Mechanical Lysis
Lysis is achieved through an optimized enzyme mix including lysozyme, mutanolysin, and proteinase K. This is followed by a short sonication pulse or mechanical shearing to ensure complete RNA release.
🧫 3. In Situ RNA Stabilization
To minimize degradation, the cells are immediately incubated with RNA preservation buffers. This step is critical for maintaining transcript integrity prior to reverse transcription.
🧪 4. RNA Capture via Artificial Polyadenylation
Bacterial mRNA is enzymatically tailed with poly(A) sequences using E. coli poly(A) polymerase. This enables downstream capture using oligo-dT primers — a crucial adaptation for compatibility with existing scRNA-seq platforms.
🔄 5. Reverse Transcription and Amplification
A modified Smart-seq2 or CEL-Seq2 method is used, often with early barcoding using unique molecular identifiers (UMIs) to reduce quantification noise.
📈 6. Library Construction and Sequencing
Libraries are prepared using low-input kits validated for short and fragmented cDNA, then sequenced on standard Illumina platforms at depths of 100,000 to 1 million reads per cell.
Advantages of the New Protocol
This bacterial scRNA-seq protocol enables key benefits for microbiology and synthetic biology labs:
✔️ Compatibility with a Wide Range of Species
Including Gram-negative species such as E. coli and Pseudomonas, as well as Gram-positive organisms like Bacillus subtilis and Streptococcus pneumoniae.
✔️ High Transcript Capture Efficiency
Yields over 60% capture rate of mRNA molecules per cell in benchmark testing — a major improvement over earlier efforts.
✔️ Accurate Gene Expression Quantification
Incorporates UMIs and quality controls for error correction, allowing reliable quantification of rare transcripts.
✔️ Integration with Multi-Omics
Can be extended to dual RNA/protein or RNA/DNA profiling for single-cell multi-modal studies in microbial systems.
Use Cases Across Scientific Domains
This protocol unlocks bacterial single-cell transcriptomics across many domains:
🧪 Environmental Microbiology
Track gene expression in environmental samples such as soil microbiomes, aquatic environments, and wastewater.
🧫 Antibiotic Resistance Studies
Analyze how different bacterial subpopulations respond to external chemical pressures and pinpoint resistant phenotypes.
🔬 Synthetic Circuit Performance
Quantify expression variability of synthetic gene circuits in individual cells during microbial fermentation or bioproduction.
🧬 Evolutionary Biology
Study population-level adaptations at the single-cell level in long-term evolution experiments.
🧾 Bioprocess Monitoring
Track stress responses and productivity in engineered microbial strains in industrial fermenters.
Studies related to these topics can be accessed through databases like PubMed Central, DOE Joint Genome Institute, and NCBI GEO.
A New Standard in Bacterial Transcriptomics
The development of a robust bacterial scRNA-seq protocol is a foundational step toward unlocking single-cell biology in prokaryotic systems. Its flexibility, efficiency, and compatibility with existing sequencing platforms make it accessible to a wide range of researchers without requiring custom-built infrastructure.
As microbial scRNA-seq becomes increasingly adopted, we anticipate a surge in discovery of new gene regulatory mechanisms, metabolic subtypes, and adaptive states that shape microbial communities across environments and industrial processes.
This approach is not just a technical enhancement—it represents a conceptual shift. It turns bacterial populations from homogenous black boxes into dynamic, trackable communities where every cell has a story.
Tags
- Biology
- LifeSciences
- Biodiversity
- Evolution
- HumanBiology
- Genetics
- MedicalBiology
- MolecularBiology
- CellBiology
- DNA
- Genomics
- Biotechnology
- Research
